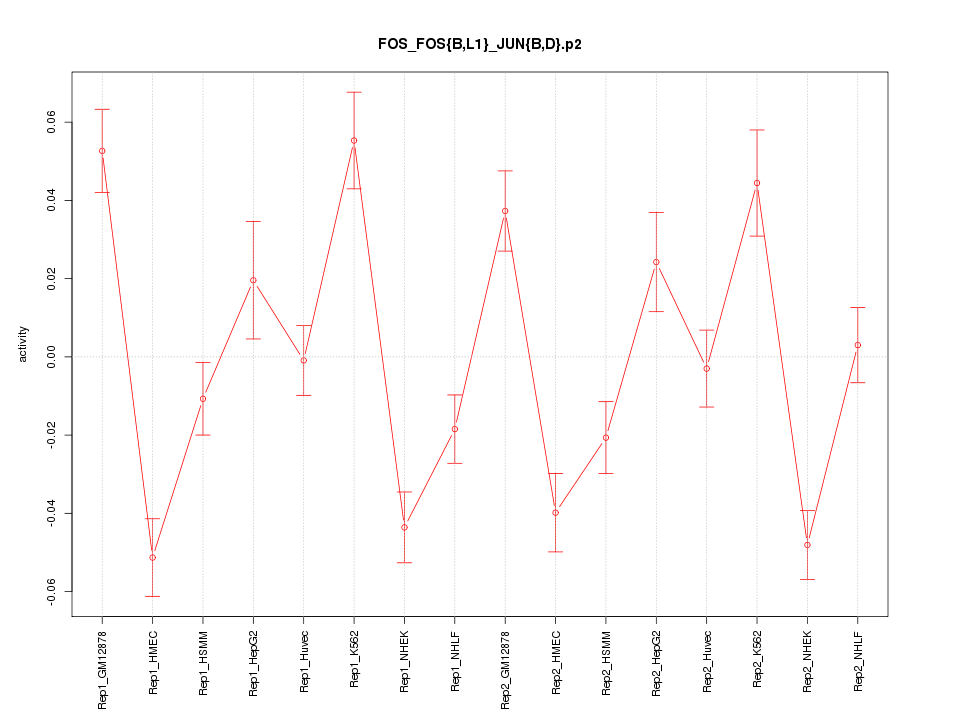
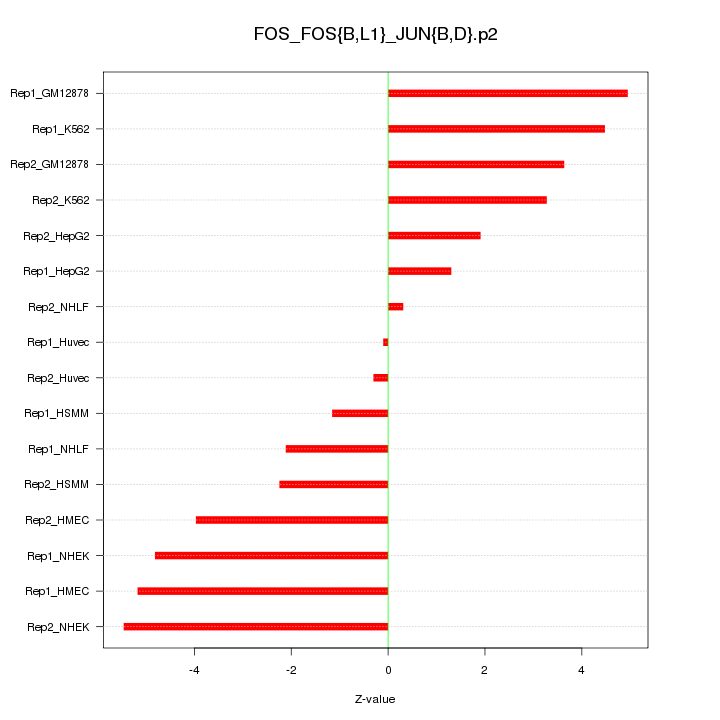
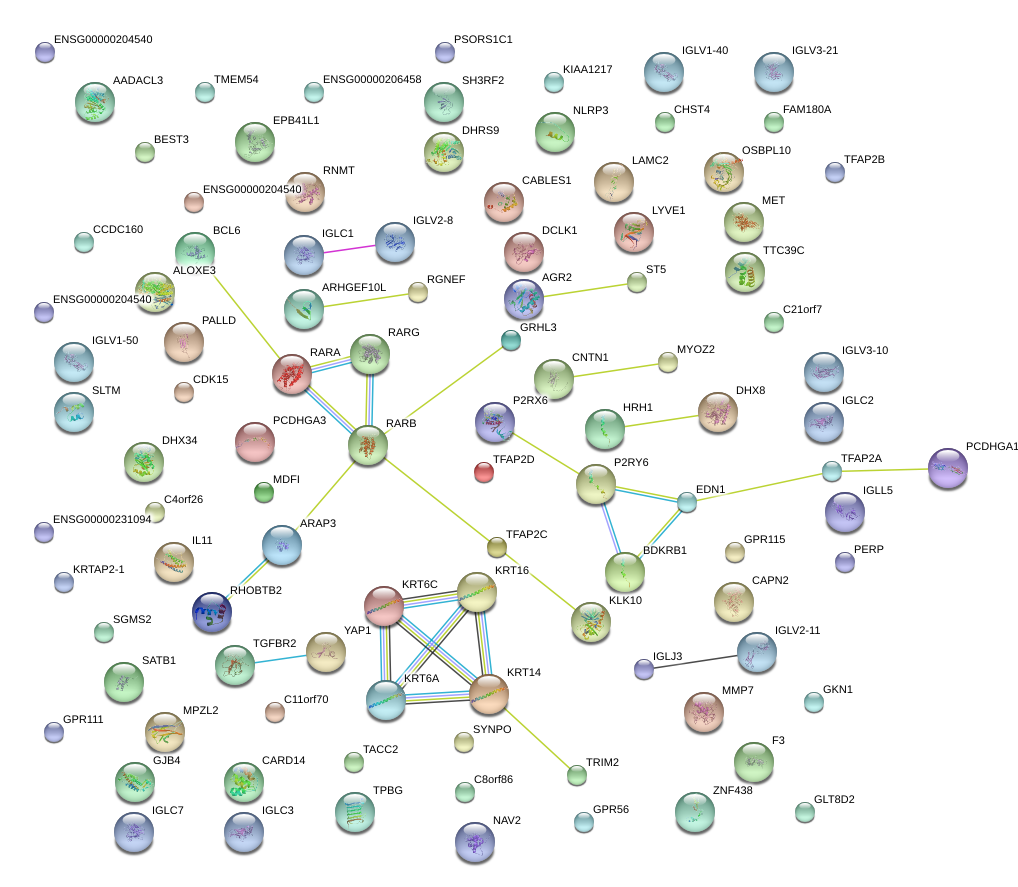
|
chr7_+_116099648
|
9.446
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr7_-_135083906
|
7.197
|
NM_205855
|
FAM180A
|
family with sequence similarity 180, member A
|
|
chr16_+_56219802
|
6.917
|
NM_001145771
NM_001145772
NM_001145773
NM_201524
NM_201525
|
GPR56
|
G protein-coupled receptor 56
|
|
chr19_-_40711034
|
6.686
|
NM_001166034
NM_001166035
NM_198538
|
SBSN
|
suprabasin
|
|
chr5_+_150000394
|
6.429
|
NM_001109974
NM_007286
|
SYNPO
|
synaptopodin
|
|
chr1_+_17719405
|
6.332
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr1_+_181421984
|
6.099
|
|
LAMC2
|
laminin, gamma 2
|
|
chr6_+_31190505
|
5.860
|
NM_014068
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1
|
|
chr1_-_94779727
|
5.505
|
NM_001178096
NM_001993
|
F3
|
coagulation factor III (thromboplastin, tissue factor)
|
|
chr2_+_169634330
|
5.384
|
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr12_-_68369322
|
5.320
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr6_-_138470160
|
5.153
|
NM_022121
|
PERP
|
PERP, TP53 apoptosis effector
|
|
chr1_+_245646080
|
4.931
|
NM_001079821
NM_001127461
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr4_+_154397975
|
4.654
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr17_-_36469869
|
4.497
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr4_+_169989680
|
4.462
|
NM_001166110
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr17_-_37022465
|
4.440
|
NM_005557
|
KRT16
|
keratin 16
|
|
chr4_+_108965167
|
4.423
|
NM_001136258
|
SGMS2
|
sphingomyelin synthase 2
|
|
chr5_+_140848905
|
4.257
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr4_+_120276386
|
4.253
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr1_+_181421796
|
4.150
|
NM_005562
NM_018891
|
LAMC2
|
laminin, gamma 2
|
|
chr11_-_101906640
|
4.092
|
NM_002423
|
MMP7
|
matrix metallopeptidase 7 (matrilysin, uterine)
|
|
chr1_+_181422016
|
3.984
|
|
LAMC2
|
laminin, gamma 2
|
|
chr19_-_60573552
|
3.961
|
NM_000641
|
IL11
|
interleukin 11
|
|
chr16_+_70117523
|
3.903
|
NM_005769
NM_001166395
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4
|
|
chr1_+_221955876
|
3.895
|
NM_001146068
|
CAPN2
|
calpain 2, (m/II) large subunit
|
|
chr6_+_47732181
|
3.837
|
NM_153839
|
GPR111
|
G protein-coupled receptor 111
|
|
chr17_+_75776193
|
3.820
|
NM_052819
|
CARD14
|
caspase recruitment domain family, member 14
|
|
chr17_-_7962531
|
3.808
|
NM_021628
|
ALOXE3
|
arachidonate lipoxygenase 3
|
|
chr8_-_38505331
|
3.763
|
NM_207412
|
C8orf86
|
chromosome 8 open reading frame 86
|
|
chr2_+_202379442
|
3.655
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr1_+_12698704
|
3.578
|
NM_001103169
NM_001103170
|
AADACL3
|
arylacetamide deacetylase-like 3
|
|
chr3_+_11153778
|
3.553
|
NM_001098213
|
HRH1
|
histamine receptor H1
|
|
chr11_+_72660894
|
3.472
|
NM_004154
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chr1_+_34997928
|
3.420
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr9_-_90456824
|
3.386
|
NM_001100111
|
LOC286238
|
hypothetical protein LOC286238
|
|
chr13_-_35327798
|
3.348
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr12_-_51173238
|
3.279
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr3_+_30622905
|
3.263
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr12_-_51912101
|
3.262
|
NM_000966
|
RARG
|
retinoic acid receptor, gamma
|
|
chr6_-_169392771
|
3.159
|
|
|
|
|
chr4_+_76700281
|
3.155
|
NM_178497
|
C4orf26
|
chromosome 4 open reading frame 26
|
|
chr10_-_21847126
|
3.089
|
|
|
|
|
chr22_+_19699441
|
3.080
|
NM_001159554
NM_005446
|
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6
|
|
chr11_-_8695974
|
3.068
|
|
ST5
|
suppression of tumorigenicity 5
|
|
chr21_+_29439768
|
3.057
|
|
C21orf7
|
chromosome 21 open reading frame 7
|
|
chr22_+_21370394
|
3.044
|
|
IGL@
|
immunoglobulin lambda locus
|
|
chr17_-_36996611
|
3.002
|
NM_000526
|
KRT14
|
keratin 14
|
|
chr6_+_47774247
|
2.981
|
NM_153838
|
GPR115
|
G protein-coupled receptor 115
|
|
chr1_-_33139425
|
2.967
|
NM_033504
|
TMEM54
|
transmembrane protein 54
|
|
chr10_-_31328426
|
2.953
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr7_-_16811131
|
2.951
|
NM_006408
|
AGR2
|
anterior gradient homolog 2 (Xenopus laevis)
|
|
chr2_+_69055208
|
2.872
|
NM_019617
|
GKN1
|
gastrokine 1
|
|
chr6_+_12398514
|
2.861
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr11_-_10546654
|
2.847
|
NM_006691
|
LYVE1
|
lymphatic vessel endothelial hyaluronan receptor 1
|
|
chr10_+_123862430
|
2.843
|
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_+_101488380
|
2.795
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr22_+_21393107
|
2.792
|
|
IGLJ3
|
immunoglobulin lambda joining 3
|
|
chr3_-_188936941
|
2.787
|
NM_001130845
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr18_+_19947314
|
2.783
|
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr3_-_31997738
|
2.771
|
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr11_+_101423378
|
2.750
|
NM_001195005
NM_032930
|
C11orf70
|
chromosome 11 open reading frame 70
|
|
chr3_+_30623376
|
2.722
|
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr6_+_41712907
|
2.692
|
|
MDFI
|
MyoD family inhibitor
|
|
chr6_+_83129641
|
2.690
|
NM_006670
|
TPBG
|
trophoblast glycoprotein
|
|
chr8_+_22900863
|
2.687
|
NM_001160036
|
RHOBTB2
|
Rho-related BTB domain containing 2
|
|
chr22_+_21464980
|
2.683
|
|
|
|
|
chr1_+_24518501
|
2.672
|
NM_001195010
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr11_+_20005704
|
2.661
|
|
NAV2
|
neuron navigator 2
|
|
chr5_+_73145098
|
2.658
|
|
RGNEF
|
190 kDa guanine nucleotide exchange factor
|
|
chr11_-_117640407
|
2.646
|
NM_005797
NM_144765
|
MPZL2
|
myelin protein zero-like 2
|
|
chr5_+_145297490
|
2.630
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr18_+_18989786
|
2.593
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr5_-_141041982
|
2.557
|
NM_022481
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr5_-_141041933
|
2.531
|
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3
|
|
chr12_-_102967828
|
2.444
|
|
GLT8D2
|
glycosyltransferase 8 domain containing 2
|
|
chr10_+_24795446
|
2.442
|
|
KIAA1217
|
KIAA1217
|
|
chr3_-_18441763
|
2.438
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr14_+_95792299
|
2.434
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr1_+_24518398
|
2.404
|
NM_198173
NM_198174
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chrX_+_133198742
|
2.331
|
NM_001101357
|
CCDC160
|
coiled-coil domain containing 160
|
|
chr20_+_34206070
|
2.323
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr6_-_10527772
|
2.306
|
NM_001042425
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr19_-_56215084
|
2.302
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr22_+_21495273
|
2.290
|
|
|
|
|
chr15_-_72952648
|
2.287
|
NM_005697
|
SCAMP2
|
secretory carrier membrane protein 2
|
|
chr3_-_31998241
|
2.275
|
NM_001174060
NM_017784
|
OSBPL10
|
oxysterol binding protein-like 10
|
|
chr10_+_24795465
|
2.273
|
|
KIAA1217
|
KIAA1217
|
|
chr5_+_145296313
|
2.273
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr20_+_44070946
|
2.252
|
NM_004994
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase)
|
|
chr9_+_123369219
|
2.248
|
NM_032552
|
DAB2IP
|
DAB2 interacting protein
|
|
chr3_-_46582806
|
2.240
|
NM_024512
|
LRRC2
|
leucine rich repeat containing 2
|
|
chr3_+_31998269
|
2.234
|
NM_001137674
|
ZNF860
|
zinc finger protein 860
|
|
chr12_-_10215942
|
2.234
|
NM_001172632
NM_001172633
NM_002543
|
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1
|
|
chr10_-_62002672
|
2.228
|
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_-_6298284
|
2.214
|
NM_145040
|
PRKCDBP
|
protein kinase C, delta binding protein
|
|
chr2_+_33213111
|
2.205
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr1_+_234372454
|
2.193
|
NM_003272
|
GPR137B
|
G protein-coupled receptor 137B
|
|
chr6_-_119137923
|
2.182
|
NM_001178035
|
C6orf204
|
chromosome 6 open reading frame 204
|
|
chr9_-_109291866
|
2.181
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr12_-_105057940
|
2.178
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr9_-_116920232
|
2.176
|
NM_002160
|
TNC
|
tenascin C
|
|
chr11_+_35596310
|
2.174
|
NM_014344
|
FJX1
|
four jointed box 1 (Drosophila)
|
|
chr4_-_89299034
|
2.165
|
NM_004827
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2
|
|
chr8_-_27528215
|
2.156
|
NM_001831
|
CLU
|
clusterin
|
|
chr20_-_43410437
|
2.145
|
NM_002999
|
SDC4
|
syndecan 4
|
|
chr1_-_84237316
|
2.126
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr7_+_22733290
|
2.113
|
NM_000600
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr6_-_150388292
|
2.077
|
NM_130900
|
RAET1L
|
retinoic acid early transcript 1L
|
|
chr1_+_150753601
|
2.067
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr9_+_137694988
|
2.059
|
NM_001001676
|
LCN9
|
lipocalin 9
|
|
chr6_+_144040751
|
2.049
|
NM_001100164
NM_001100165
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr17_-_36928604
|
2.048
|
NM_002275
|
KRT15
|
keratin 15
|
|
chr4_+_40032146
|
2.041
|
NM_017581
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9
|
|
chr2_+_121271336
|
2.035
|
NM_005270
|
GLI2
|
GLI family zinc finger 2
|
|
chr22_+_21053972
|
2.030
|
|
|
|
|
chr18_-_72336133
|
2.027
|
NM_014643
|
ZNF516
|
zinc finger protein 516
|
|
chr3_-_152517204
|
2.026
|
NM_023915
|
GPR87
|
G protein-coupled receptor 87
|
|
chr2_-_69033515
|
1.992
|
NM_182536
|
GKN2
|
gastrokine 2
|
|
chr1_+_34993234
|
1.972
|
NM_005268
|
GJB5
|
gap junction protein, beta 5, 31.1kDa
|
|
chr1_-_151280201
|
1.960
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr5_+_36642424
|
1.954
|
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr6_+_107066422
|
1.950
|
NM_001624
|
AIM1
|
absent in melanoma 1
|
|
chr5_-_137099361
|
1.947
|
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr19_-_56164628
|
1.946
|
NM_002774
|
KLK6
|
kallikrein-related peptidase 6
|
|
chr6_+_45404031
|
1.937
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr2_+_220200530
|
1.926
|
NM_005070
NM_201574
|
SLC4A3
|
solute carrier family 4, anion exchanger, member 3
|
|
chr8_-_27528105
|
1.924
|
|
CLU
|
clusterin
|
|
chr2_+_113591940
|
1.915
|
NM_000577
NM_173841
NM_173843
|
IL1RN
|
interleukin 1 receptor antagonist
|
|
chr14_+_62740878
|
1.894
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr5_+_36644187
|
1.870
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_-_150398327
|
1.869
|
NM_001122965
|
RPTN
|
repetin
|
|
chr18_+_19706979
|
1.853
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr9_+_676640
|
1.848
|
|
|
|
|
chr11_-_119499188
|
1.832
|
|
TRIM29
|
tripartite motif containing 29
|
|
chr11_-_2907169
|
1.829
|
NM_003311
|
PHLDA2
|
pleckstrin homology-like domain, family A, member 2
|
|
chr5_-_137099602
|
1.825
|
NM_017415
|
KLHL3
|
kelch-like 3 (Drosophila)
|
|
chr6_+_1556543
|
1.823
|
|
FOXC1
|
forkhead box C1
|
|
chr7_-_142816019
|
1.817
|
NM_005232
|
EPHA1
|
EPH receptor A1
|
|
chr22_+_20899155
|
1.809
|
|
|
|
|
chr11_-_117640166
|
1.808
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr7_-_107430566
|
1.794
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr12_+_13240868
|
1.778
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_+_24522116
|
1.775
|
NM_021180
|
GRHL3
|
grainyhead-like 3 (Drosophila)
|
|
chr1_-_7923464
|
1.752
|
NM_001561
|
TNFRSF9
|
tumor necrosis factor receptor superfamily, member 9
|
|
chr17_-_51155073
|
1.749
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr11_-_76676110
|
1.738
|
NM_182833
|
GDPD4
|
glycerophosphodiester phosphodiesterase domain containing 4
|
|
chr17_-_37034288
|
1.733
|
NM_000422
|
KRT17
|
keratin 17
|
|
chr6_+_106653429
|
1.728
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr1_+_27591738
|
1.721
|
NM_005281
|
GPR3
|
G protein-coupled receptor 3
|
|
chr17_+_71229110
|
1.717
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr14_-_22658078
|
1.716
|
NM_001805
|
CEBPE
|
CCAAT/enhancer binding protein (C/EBP), epsilon
|
|
chr2_+_233605625
|
1.703
|
NM_005383
|
NEU2
|
sialidase 2 (cytosolic sialidase)
|
|
chr18_-_26996742
|
1.700
|
NM_004948
NM_024421
|
DSC1
|
desmocollin 1
|
|
chr15_+_99276982
|
1.698
|
NM_024652
|
LRRK1
|
leucine-rich repeat kinase 1
|
|
chr12_-_10173947
|
1.695
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr12_+_13240983
|
1.695
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_+_151270301
|
1.689
|
NM_003125
|
SPRR1B
|
small proline-rich protein 1B
|
|
chr19_-_40684638
|
1.671
|
NM_001035516
|
DMKN
|
dermokine
|
|
chr12_+_52733927
|
1.660
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr8_-_68821131
|
1.659
|
NM_020361
|
CPA6
|
carboxypeptidase A6
|
|
chr5_-_54317102
|
1.652
|
NM_001135604
NM_007036
|
ESM1
|
endothelial cell-specific molecule 1
|
|
chr1_-_85816520
|
1.652
|
NM_001134445
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr4_-_152366970
|
1.646
|
NM_001009555
NM_001128923
|
SH3D19
|
SH3 domain containing 19
|
|
chr12_+_13240987
|
1.641
|
|
EMP1
|
epithelial membrane protein 1
|
|
chr1_+_19842309
|
1.637
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr14_-_104507862
|
1.636
|
|
AHNAK2
|
AHNAK nucleoprotein 2
|
|
chr19_-_48661563
|
1.627
|
NM_014400
|
LYPD3
|
LY6/PLAUR domain containing 3
|
|
chr17_-_36409632
|
1.625
|
NM_031959
|
KRTAP3-2
|
keratin associated protein 3-2
|
|
chr2_+_27573206
|
1.620
|
NM_001486
|
GCKR
|
glucokinase (hexokinase 4) regulator
|
|
chr2_+_173394560
|
1.618
|
NM_001100397
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr11_-_117640196
|
1.617
|
|
MPZL2
|
myelin protein zero-like 2
|
|
chr6_+_121798443
|
1.616
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr3_-_71857097
|
1.608
|
NM_001134651
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr2_+_135312655
|
1.604
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chr19_+_40895669
|
1.590
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr18_+_50646616
|
1.588
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr1_+_103898844
|
1.577
|
NM_020978
|
AMY2B
|
amylase, alpha 2B (pancreatic)
|
|
chr17_+_55269974
|
1.575
|
|
|
|
|
chr1_-_17179694
|
1.567
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr11_-_66898223
|
1.562
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr16_+_4546491
|
1.561
|
NM_001145011
|
LOC342346
|
hypothetical protein LOC342346
|
|
chr16_+_88515917
|
1.558
|
NM_001197181
|
TUBB3
|
tubulin, beta 3
|
|
chr8_-_125252884
|
1.558
|
|
C8orf78
|
chromosome 8 open reading frame 78
|
|
chr17_+_46067114
|
1.555
|
NM_001144070
NM_003786
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3
|
|
chr19_-_56204615
|
1.547
|
NM_012315
|
KLK9
|
kallikrein-related peptidase 9
|
|
chr2_+_36777336
|
1.544
|
NM_001177969
NM_001177970
NM_001177971
NM_001177972
NM_053276
|
VIT
|
vitrin
|
|
chr6_-_31756119
|
1.542
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr1_-_221920023
|
1.541
|
NM_001143962
|
CAPN8
|
calpain 8
|
|
chr19_+_43572679
|
1.534
|
NM_001039616
NM_001042522
|
SPRED3
|
sprouty-related, EVH1 domain containing 3
|
|
chr9_+_131974506
|
1.532
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr14_-_22515725
|
1.528
|
|
JUB
|
jub, ajuba homolog (Xenopus laevis)
|
|
chr1_-_120112989
|
1.525
|
NM_001166107
NM_005518
|
HMGCS2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 (mitochondrial)
|
|
chr2_-_166359016
|
1.507
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr1_+_35019376
|
1.499
|
NM_024009
|
GJB3
|
gap junction protein, beta 3, 31kDa
|
|
chr7_-_93357881
|
1.499
|
NM_006528
|
TFPI2
|
tissue factor pathway inhibitor 2
|
|
chr12_-_121224676
|
1.490
|
NM_001014336
|
IL31
|
interleukin 31
|
|
chr22_+_21042091
|
1.489
|
|
IGLV1-44
IGL@
|
immunoglobulin lambda variable 1-44
immunoglobulin lambda locus
|
|
chr1_+_85300580
|
1.480
|
NM_145172
|
WDR63
|
WD repeat domain 63
|
|
chr6_+_237033
|
1.462
|
NM_020185
|
DUSP22
|
dual specificity phosphatase 22
|